日本医科大学、医学部、生化学・分子生物学(代謝・栄養学)所属の 岩崎 俊雄グループでは、好熱菌等を主材料とした各種金属酵素の構造機能進化や、アミノ酸要求性の新規大腸菌発現宿主株作成につき、国内外の様々な研究機関と共同研究を実施しています。
Microbial "mitoNEET" homologs
The mammalian mitochondrial outer membrane protein, named "mitoNEET" [on the basis of its subcellular location on the outer membrane of mitochondria and the internal amino-acid sequence stretch of the mouse and human proteins, Asn-Glu-Glu-Thr (NEET)], was identified as a possible target protein of pioglitazone by cross-linking with a photoaffinity probe by Colca et al. The drug pioglitazone is a member of the thiazolidinedione class of insulin sensitizers for the treatment of type II diabetes, a complex metabolic disease of prevailing public health concern that is characterized in the initial stage by insulin resistance. Deficiency of mitoNEET expression in mice results in a compromise in the respiratory capacity of cardiac mitochondria.
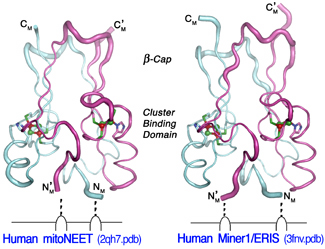
Recent crystallographic studies on the recombinant soluble (CDGSH-type zinc-finger-like) domain of human mitoNEET at 1.4-1.8 Å resolution (PDB codes: 2qd0, 2r13, 2qh7 and 3ew0) have revealed that the recombinant mitoNEET soluble domain is a homodimer with each subunit binding a unique [2Fe–2S] cluster (but no Zn2+) in a three-cysteine plus one-histidine ligand environment. A very similar mammalian protein called Miner1/ERIS/CDGSH2, originally annotated as a CDGSH-type zinc-finger protein of unknown function, is also a member of the mitoNEET-like [2Fe-2S](Cys)3(His)1 protein superfamily, and localizes to both the outer membrane of mitochondria and the endoplasmic reticulum (with a 6:1 ratio). Mis-splicing of the mRNA that encodes this Miner1/ERIS protein is causative in the genetic disease Wolfram Syndrome 2 (WFS2). Knock-out mice are also symptomatic, with similar signs of early aging and optical atrophy.
Despite the potential importance in pharmacology and clinical medicine, the in vivo function of the mitoNEET protein superfamily remains elusive. We are interested in the structure and (redox) function of this unique iron-sulfur protein superfamily.
up
Biophysical studies on rat mitoNEET: spin-spin interactions between two [2Fe-2S] clusters
Interest in the redox chemistry and physiology of the mitoNEET superfamily has been heightened by the recent availability of crystal structures (Fig. 1). However, information about the electronic structure and interaction of this novel "twin" [2Fe-2S] cluster system was very limited, and the site of iron reduction and the cluster's magnetic interaction with the protein environment which account for the electron transfer function and mechanism remained elusive in the mammalian mitoNEET system.
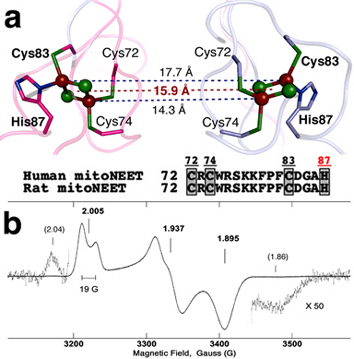
Figure 1. (a) Relative location of two clusters in the human mitoNEET (soluble domain) dimeric structure (2QH7.pdb). In this structure, the outermost iron is coordinated by solvent-exposed His87 and Cys83 ligands, and the innermostiron is coordinated by Cys74 and Cys72 ligands. (b) X-band continuous-wave (CW) electron paramagnetic resonance (EPR) spectrum of reduced rat mitoNEET soluble domain at 20 K. Additional weak wings, probably resulting from magnetic interaction of the reduced clusters, accompany the main EPR signal on the high and low field sides. Microwave frequency and power, 9.037 GHz and 0.01 mW; sample concentration, ~4 mM (per protomer).
Strong antiferromagnetic coupling between the electron spins of two irons via a bridging structure produces an EPR-silent (S=0) ground state in the oxidized Fe3+-Fe3+ form of the [2Fe-2S] cluster and a paramagnetic S=1/2 ground state in the reduced Fe3+-Fe2+ form. Thus, the reduced [2Fe-2S] cluster in frozen solution is characterized by the anisotropic EPR spectrum, typically as a result of a rhombic g-tensor. The distinguishing feature between the reduced plant ferredoxin [2Fe-2S](Cys)4 and Rieske [2Fe-2S](Cys)2(His)2 clusters is the average value of the g-tensor components gav, which is ~1.96 for plant ferredoxins and ~1.91 for Rieske proteins. The cluster in the mitoNEET protein family is the first example of a "redox-active" [2Fe-2S] cluster with asymmetrical coordination of one of the irons (Fig. 1). Before the discovery of the mitoNEET protein family, we had rationally engineered a [2Fe-2S](Cys)3(His)1 environment into the hyperthermophilic archaeal Rieske-type ferredoxin (ARF) scaffold of Sulfolobus solfataricus (DDBJ-EMBL-GenBank code, AB047031) with replacement of the His64→Cys ligand (H64C-ARF). This accommodated a fairly thermostable [2Fe-2S] cluster in the oxidized form. However, its high sensitivity to dithionite reduction even under anaerobic conditions (leading to irreversible reductive cluster breakdown) precluded detailed characterization of its reduced state. Thus, the mammalian mitoNEET system opens a new path to explore a deeper analysis of the paramagnetic form of a unique (and redox-active) [2Fe-2S](Cys)3(His)1 cluster.
Continuous-wave (CW) X-band EPR spectrum of the dithionite-reduced, the rat mitoNEET soluble domain (residues 32-108; NCBI_GeneID code 294362), containing two nominally identical [2Fe-2S](Cys)3(His)1 clusters per dimer, possesses an anisotropic lineshape with a width of ~250 G, corresponding to rhombic g-tensor (Fig. 1b). The principal values of the g-tensor can be estimated by the values gz=2.005, gy=1.937, and gx=1.895. The average value of these components (gav ~1.945) is reasonably between the values of the plant ferredoxin (~1.96) and Rieske (~1.91) [2Fe-2S] clusters. In addition, EPR spectrum shows the splitting ~19 G of the gz component in the low field of the spectrum and a broadening of the central gy component from the poorly resolved splitting of the order ~10 G (Fig. 1b). These features resulted from the interaction of the electron spins of two adjacent, reduced clusters in the dimeric unit (Fig. 1). This interaction was not recognized in the previously published spectrum of the reduced mitoNEET soluble domain.
X-ray crystal structure of the human mitoNEET soluble domain gives distances of 14.3 Å and 17.7 Å between the innermost and outermost iron pairs of two [2Fe-2S] clusters, respectively (Fig. 1a). The distance between the geometrical centers of the two clusters is 15.9 Å. The parameter D=g^2β^2/r^3 =18500/r^3 (D in Gauss, r in Å), which characterizes an order of spin-spin interaction between two reduced clusters in the approximation considering them as two S=1/2 dipoles, varies within the interval 7.7 and 3.3 G for these distances (4.6 G for the center). Thus, the observed large gz splitting ~19 G (Fig. 1b) cannot be explained by the simple dipole-dipole interaction of two S=1/2 spins separated by the reported crystallographic distance (~14-18 Å) of the two adjacent clusters (Fig. 1a).
Our calculations demonstrate that the observed gz splitting in the EPR spectrum of the reduced rat mitoNEET is more adequately described by employing the “local spin model” which considers the spin-spin interactions between all four irons in reduced clusters (Fig. 2a). This model favors the reduction of the outermost iron with His87 and Cys83 ligands (Fig. 2b). These peculiarities suggest that the mammalian mitoNEET might have evolved to conduct rapid intramolecular electron transfer reaction in the dimeric unit.
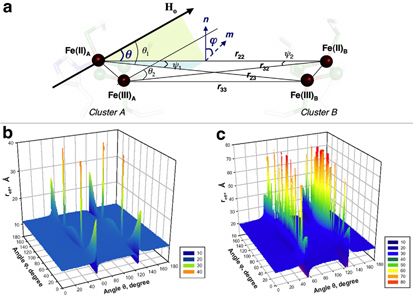
Figure 2. Schematic presentation of vectors and angles for the qualitative calculation by employing the local spin model (a). In this figure, the r22 and r33 form the same angle θ with the external magnetic field vector Ho. ϕ is the angle between the planes containing the vectors (r23,r22) and (r22,Ho) and is equal to that between their normals n (to the plane containing r23 and r22) and m (to the plane containing r22 and Ho). The deviation of m from n depends on the orientation of Ho. In panels (b) and (c) prepared in 3D presentation by Prof. Sergei Dikanov (University of Illinois at Urbana-Champaign), the effective distance reff (Å) was calculated as a function of angles θ and ϕ (with the step 1°) when the outermost iron pair (reff ~11 Å for most orientations of the magnetic field relative to the molecular axes of the cluster defined by angles θ and ϕ) (b) or the innermost iron pair (reff ~22 Å, which is substantially longer) (c) undergoes the reduction in rat mitoNEET. Further details are given in the original article of this work.
2D pulsed EPR (HYSCORE) characterization of rat mitoNEET
Reduction of the outermost iron pair with His87 and Cys83 ligands in the mixed-valent state of the mitoNEET [2Fe-2S] cluster, indicated by local spin model analysis (Fig. 2), can be addressed independently by the two-dimensional pulsed EPR (also called hyperfine sublevel correlation, HYSCORE) characterization of hyperfine couplings from directly coordinated nitrogen of the His87 ligand. The representative 14N HYSCORE spectrum of the reduced 14N(N/A)-rat mitoNEET is shown in Fig. 3a. It consists of two quadrants containing cross-peaks from several nitrogens. Previous analysis with different values of 14N hyperfine couplings allows us to assign extended cross-peaks in the (+–) quadrant to strongly coupled (coordinated) 14Nδ of the histidine ligand (His87), and two pairs of cross-peaks in the (++) quadrant with an approximately circular shape of small radius to other, non-liganding nitrogens. The 14Nδ nuclear frequencies from the His87 ligand of rat mitoNEET, determined from the 14N HYSCORE spectra recorded at the fields corresponding to principal values of the g-tensor, provide an estimate of diagonal components of the hyperfine and quadrupole tensors in the g-tensor coordinates and the isotropic hyperfine constant (14)a=6.1 MHz.
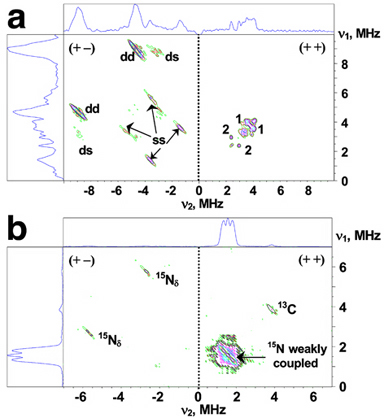
Figure 3. HYSCORE spectra in contour presentation of the reduced [2Fe-2S] cluster in 14N(N/A, 99.63%) (a) and uniformly 15N-labeled (b) rat mitoNEET soluble domain; dd, ds, and ss are cross-peaks correlating d, double-quantum, and s, single-quantum transitions. Magnetic field, time τ, and microwave frequency were: 3672 G, 136 ns, 9.706 GHz (a); 3580 G, 136 ns, 9.704 GHz (b).
The 15N HYSCORE spectrum of the uniformly 15N-labeled rat mitoNEET (Fig. 3b) shows only one pair of cross-peaks in the (+−) quadrant from the His87 15Nδ (nuclear spin I=1/2, no quadrupole moment). The isotropic and anisotropic parts of axial hyperfine tensor (15)a=8 MHz, (15)T=1.1 MHz (corresponding to (14)a=5.7 MHz and (14)T=0.8 MHz) defining principal values of the tensor were obtained by the contour lineshape analysis (data not shown). Notably, these parameters for His87 Nδ in rat mitoNEET are only ~10% above the corresponding values for one of the two Nδ(His) 's (with larger hyperfine tensor) in Rieske-type proteins (see also the original article of this work). In the Rieske-type protein system, the Nδ(His) hyperfine tensors depend on anisotropic interaction with the electron spins of the Fe(II) and Fe(III), and the spin density transferred onto the strongly coupled (i.e., coordinated) 14Nδ(His) atom. The constrains on Nδ(His) location (stable Fe-Nδ(His) distances and angles between Fe-Fe and Fe-Nδ(His)) based on the available crystal structures would account for the similarity of hyperfine tensors between different proteins, if they are determined mainly by the location of the Nδ(His) nuclei relative to the electron spins of the cluster, and are relatively insensitive to variations in orientation of the imidazole plane. The latter influences only the overlap of the iron and nitrogen orbitals responsible for the spin density transfer onto the ligand. This means that the hyperfine tensor of the strongly coupled Nδ(His) and its principal directions mainly reflect the position of the Nδ(His) atom itself relative to the iron-sulfur cluster, and therefore would not provide direct information about changes in orientation of the imidazole plane, so long as the Nδ(His) location remains approximately constant. Thus, the HYSCORE analysis provides the second indication that the Fe2+ site in the reduced cluster of rat mitoNEET is the outermost iron.
It should be added that maximum hyperfine and quadrupolar splittings from 14Nδ(His87) in rat mitoNEET are observed along gx(min) axis, in contrast to the gz(max) axis for both 14Nδ(His) ligands in Rieske-type proteins. The hyperfine tensor of the directly coordinated Nδ(His) and its principal directions mainly reflect the position of the nitrogen atom itself relative to the iron-sulfur cluster, whereas the principal directions of the nuclear quadrupole interaction tensor are determined by the geometry of the electron orbitals around the nucleus and are associated with the ligand molecule itself. Both mitoNEET- and Rieske-type cluster systems have one common Nδ(His) ligand located at the equivalent position relative to the [2Fe-2S] cluster (as judged by similar Fe-Nδ(His) distances, angles between the Fe-Fe and Fe-Nδ(His) and orientations of imidazole plane), which would account for the similar hyperfine and quadrupole tensors (see the original article of this work). Based on these considerations, it seems reasonable to postulate a possible difference in orientation of the gmax, gmid, and gmin principal axes of the g-tensors relative to the basic molecular axes between the mitoNEET and Rieske cluster systems.
up
14N and 15N HYSCORE characterization of rat mitoNEET also suggested marked differences between the mitoNEET and Rieske [2Fe-2S] protein systems in regard to the cluster's magnetic interaction with the protein environment (Figs. 4-6). In contrast to Rieske proteins, we did not detect (non-coordinated) peptide nitrogen (Np(s)) with hyperfine couplings significantly exceeding the Nε(His) coupling. It is likely that peptide nitrogens involved in hydrogen-bond formation with the reduced cluster of the mammalian mitoNEET soluble domain carry less spin density than those of Rieske-type proteins, reflecting their structural differences, including configurations of the liganding residues to the reduced cluster. In conjunction with the marked instability of a reduced [2Fe-2S](Cys)3(His)1 cluster rationally engineered into the archaeal Rieske-type ferredoxin scaffold, our results may imply the importance of the cluster's interaction with the protein environment for the assurance of biological redox function. Although the extensive nuclear magnetic resonance (NMR) studies on bacterial rubredoxin have revealed some quantitative relations involving unpaired spin density onto peptide nitrogens, strength of hydrogen bonds, and redox properties, NMR identification and full assignment of resonances from a paramagnetic center of a polynuclear iron-sulfur protein with slow electronic relaxation rates (e.g. the [2Fe-2S](Cys)4 ferredoxins) are still challenging. Because up to now the parametrization of weakly coupled subsystems has not yet been optimized at the density functional theory level, the present results can also be used in theoretical analysis for selection of an appropriate model of the mixed-valence state of the mammalian mitoNEET system.
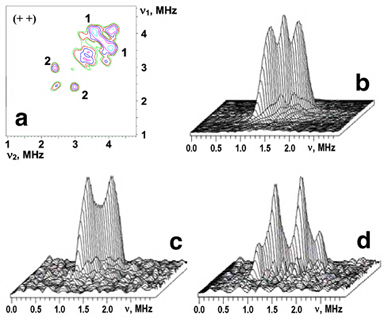
Figure 4. The (++) quadrant of the 14N HYSCORE spectrum taken from Fig. 3a (a); 3D stacked presentation of the line from weakly coupled 15N nuclei in the (++) quadrant of 15N HYSCORE spectrum (recorded near gy) from Fig. 3b (b); the same as (b) for uniformly 15N-labeled, reduced rat mitoNEET (c) and a low-potential, archaeal Rieske-type ferredoxin (ARF) from S. solfataricus (d). Both (c) and (d) were recorded near gz area of the EPR line. Magnetic field, time τ, and microwave frequency, respectively: 3450 G, 136 ns, 9.704 GHz (c); 3425 G, 136 ns, 9.690 GHz (d). Notably, the intensity of the central “matrix” peak at the diagonal point (15νN,15νN) is suppressed in (c) and (d) in contrast with (b).
Figure 5. 3D-stacked presentation of the HYSCORE spectra in the (++) quadrant of 14N(N/A)-rat mitoNEET (a) and partially and selectively 15N(3)-His-labeled rat mitoNEET (b). The two-pulse ESEEM amplitude of the 15N(3)-His-labeled rec mitoNEET (b) showed ~30% substitution of the coordinated His87 14Nδ by 15N (not shown). In the (++) quadrant of the HYSCORE spectra, this sample shows a new doublet (15N) located symmetrically around diagonal point (15νN,15νN) with the splitting ~0.45-0.55 MHz [and varying width up to 0.2 MHz due to difference in contributing orientations] (b). The appearance of this doublet is accompanied by ~30% decrease of the relative intensity of the cross-peaks 2 relative to the cross-peaks 1 (indicated by arrows in (b)). Spectra were recorded at similar conditions with magnetic field 3620 G, time τ=136 ns, microwave frequency ~9.70 GHz.
Figure 6. 3D presentation of the HYSCORE spectra in the (++) quadrant of the partially and selectively 15N(3)-His-labeled rat mitoNEET soluble domain recorded near gx (a) and gy (c), and superimposition of the 3D plots for the uniformly 15N-labeled rat mitoNEET (blue) and the difference [uniformly 15N-labeled minus 15N(3)-His-labeled rat mitoNEET multiplied by coefficient 3.5 (i.e., corrected for ~30% substitution efficiency for 15N(3)-His), see Fig. 5] spectra (red) near gx (b) and gy (d). The difference spectra (b,d) indicate ~25% contribution of His87 15Nε to the amplitude of major doublet with the splitting ~0.4-0.5 MHz in gx (b) and gz (not shown) area and less than ~10% contribution in gy (d) area. This experiment also indicates that scrambling of randomly 15N-labeled peptide and other side chain nitrogens around the cluster does not occur with this partially 15N(3)-His-labeled sample (a,c), which would contribute to the central diagonal point (15νN,15νN) and to areas around this peak in the HYSCORE spectra [as seen with the uniformly 15N-labeled protein (b,d)]. Spectra were recorded at similar conditions with magnetic field 3670 G (near gx) for (a,b), and 3580 G (near gy) for (c,d); time τ=136 ns; microwave frequency ~9.70 GHz.
*One can follow the link to read the original article of this work.
up
Crystallization of TthNEET0026
Thermus thermophilus strain HB8 (JCM10941) is a Gram-negative thermophilic bacterium isolated from the Mine hot springs, Izu, Japan. The whole genomic DNA sequence has been analyzed (http://www.thermus.org) and genetic manipulation systems for the introduction, disruption and homologous expression of intrinsic/foreign genes have been developed. A homology search of the deduced human and rat mitoNEET sequences against the complete T. thermophilus HB8 genome database using the NCBI BLAST and Kyoto Encyclopedia of Genes and Genomes (KEGG) servers indicated at least two hypothetical genes, ttha0026 and ttha1309, that code for mitoNEET-like soluble proteins (our unpublished results). The ttha1309 gene is tentatively annotated as a short antiframe (ORF) located within a hypothetical gene cluster coding for a putative radical SAM enzyme family (data not shown). The ttha0026 gene is a part of the (hypothetical) ttha0025–ttha0026 cluster coding for a putative leucine-rich membrane protein (TTHA0025; 157 amino acids) and a mitoNEET-like water-soluble protein (TTHA0026; 69 amino acids) (Fig. 7). Moreover, the four ligand residues (Cys37, Cys39, Cys48 and His52) in the [2Fe–2S] cluster-binding motif of human mitoNEET are strictly conserved in the TTHA0026 protein (Fig. 7), even though TTHA0026 is tentatively categorized as a hypothetical CDGSH-type zinc-finger protein. It is therefore of particular interest to investigate whether TTHA0026 is indeed a bacterial prototypal model of mitoNEET carrying a mitoNEET-like [2Fe–2S] cluster and to address its functionalities through elucidation of its structure at atomic resolution and analyses of the phenotypes and the in vivo network of genetically manipulated T. thermophilus strains.
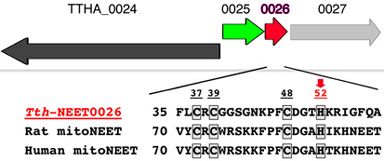
Figure 7. The Schematic organization of the ttha0025–ttha0026 gene cluster and its adjacent regions in the genomic DNA sequence of T. thermophilus HB8. There is no direct information available concerning the specificfunction of the products of the ttha0024–ttha0027 genes [TTHA0025 is a putative (leucine-rich) membrane protein, TTHA0026 is a water-soluble homologue of the mammalian mitoNEET soluble domain (TthNEET0026) and TTHA0027 is a putative potassium channel β-subunit homologue]. Multiple sequence alignment of selected mitoNEET-like proteins indicates the conservation of one histidine (indicated by a red arrow) and three cysteine residues, which provide the [2Fe–2S](Cys)3(His)1 cluster-binding motif (boxed) in the recombinant human mitoNEET soluble-domain fragment. The overall sequence identity between TthNEET0026 (69 amino acids) and the human mitoNEET soluble domain of known structure (76 amino acids) is 20.3%. Accession numbers: T. thermophilus HB8 TthNEET0026, ORFannotation/KEGG code TTHA0026, NCBI GeneID code 3168947; Rattus norvegicus (rat) mitoNEET (Cisd1), NCBI GeneID/KEGG code 294362; Homo sapiens (human) mitoNEET soluble-domain fragment, PDB codes 2qd0, 2r13 and 2qh7.
The thermophilic homolog of mitoNEET (TTHA0026) from T. thermophilus HB8 was heterologously overproduced in E. coli BL21-CodonPlus(DE3)-RIL strain and purified as a water-soluble prototypal protein containing the mitoNEET-like [2Fe-2S] cluster (Fig. 8). These results indicate the presence of the redox-active, mitoNEET-like dimeric [2Fe-2S](Cys)3(His)1 protein family across various organisms from thermophiles to mammals. The resultant recombinant protein, named TthNEET0026, has been crystallized in its oxidized form by the hanging-drop vapor diffusion method using 17%(w/v) polyethylene glycol 4000, 8.5%(v/v) 2-propanol, 15%(v/v) glycerol and 85 mM HEPES–NaOH buffer, pH 7.2 (Fig. 8). The dark reddish crystals diffract to 1.80 Å resolution and belong to the hexagonal space group P43212, with unit-cell parameters a = 45.51, c = 84.26 Å. The asymmetric unit contains one protein molecule.
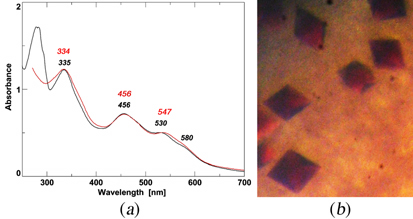
Figure 8. (a) The visible–UVabsorption spectra of purified TthNEET0026 (red trace) and rat mitoNEET [2Fe–2S] cluster-binding domain (residues 32–108; black trace). The absorption maxima (nm) are indicated in the figure. (b) Typical crystals of recombinant TthNEET0026. The maximum dimensions of the crystals are approximately 0.1 X 0.1 X 0.15 mm.
*One can follow the link to read the original article of this work.
up
Unpaired electron spin density distribution across reduced [2Fe-2S](His)n(Cys)4-n (n = 0, 1, 2) cluster ligands by 13Cβ-cysteine labeling
The biological functions of Fe-S clusters are predominantly determined by the number and types of coordinating ligands (typically cysteine and histidine) that modifiy the electronic structure of the cluster, and electron transfer between redox-active cofactors and their biological partners depends critically on the extent of electron orbital overlap. The proximity of the redox centers is the primary factor in facilitating efficient electron transfer, which can be modeled by the edge-to-edge distance between the cofactors, or by more sophisticated models using the distribution of electron spin density as a probe in quantum/molecular mechanics calculations for mapping out potential electron transfer pathways. The distribution of electrons within the redox-active cofactor is directly linked to the biological electron transfer mechanism, where the pattern of unpaired spin delocalization may play an important role.
In an orientation-selective pulsed EPR experiment, the hyperfine interactions of the paramagnetic center with the surrounding nuclei are measured as a function of the g-tensor orientation. This is achieved by taking measurements at different magnetic fields, thereby exciting only a subset of g-values out of the full g-tensor at a time. In order to determine the hyperfine interactions with the nearby Cys-13Cβ's, the orientation-selective 13C-HYSCORE analysis is performed on selectively Cys-13Cβ-labeled [2Fe-2S](His)n(Cys)4−n (n = 0, 1, 2) protein samples (reduced in the S = 1/2 spin state) in the 15N-protein background, prepared by using E. coli cysteine auxotrophic expression host strain YM154 (Figs. 9, 10).
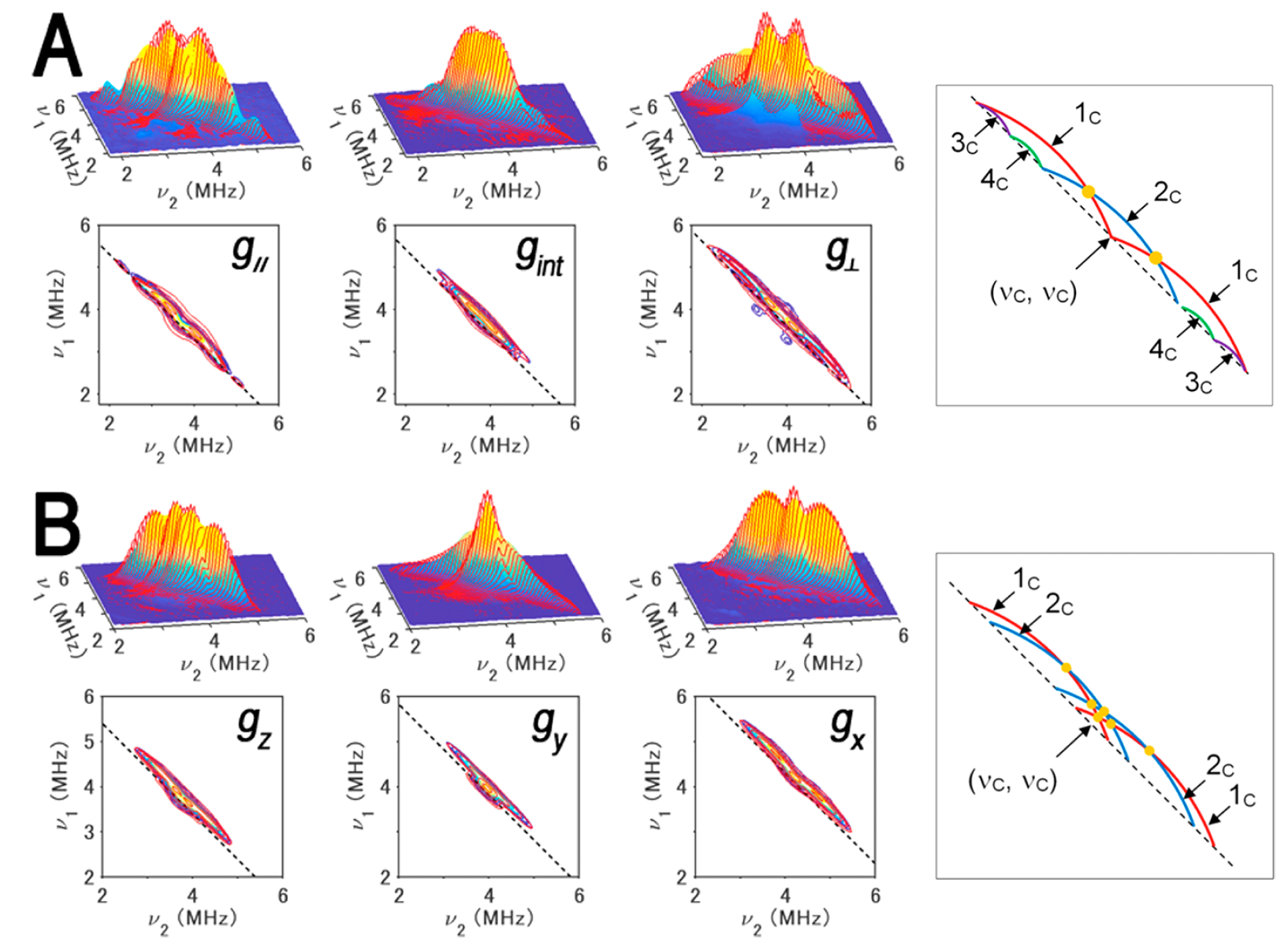
Figure 9. 13Cβ(Cys) HYSCORE spectra of FdxB measured at g|| (left), gint (middle left), and g⊥ (middle right) (A) and ARF measured at gz (gmax) (left), gy (gint) (middle left), and gx (gmin) (middle right) (B), shown in three-dimensionally stacked (top) and contour (bottom) representations, respectively, and schematic diagrams (right) of the powder 13C HYSCORE spectra of 13Cβ-Cys-labeled, reduced FdxB (A) and ARF (B) in the 15Nprotein background for the simulated cross-ridge line shapes (the schematic figures are not drawn to scale, and the cross-ridge curvature is exaggerated for the sake of clarity). The spectral simulations are colored red overlaying the experimental 13C HYSCORE spectra in the (++) quadrant. In the contour representations, the dashed lines along the antidiagonal are defined by να + νβ = 2ν(13C). In the optimized EasySpin simulations of the FdxB spectra (A), two 13Cβ(Cys) tensors, 3c (a = 2.8 ± 0.1 MHz; T = 0.5 ± 0.1 MHz; δ = 0; Euler angles α = 0°, β = 50°, and γ = 0°) and 4c (a = 1.8 ± 0.2 MHz; T = 0.5 ± 0.2 MHz; δ = 0; Euler angles α = 0°, β = −60°, and γ = 0°), have simple cross-peak shapes lying along the dashed antidiagonal line defined by να + νβ = 2ν(13C), and two anisotropic 13Cβ(Cys) tensors of similar low rhombicity, 1c (a = 1.1 ± 0.2 MHz; T = 1.3 ± 0.1 MHz; δ = 0.2 ± 0.1; Euler angles α = 40 ± 180°, β = 81 ± 12°, and γ = 33 ± 62°) and 2c (a = −0.2 ± 0.2 MHz; T = 1.2 ± 0.1 MHz; δ = 0.2 ± 0.2; Euler angles α = −30 ± 180°, β = −19 ± 11°, and γ = 82 ± 180°), make up the more intensive “arc-shaped” cross-ridges shifted off of the antidiagonal in the (++) quadrant (right), where the principal values of the rhombic hyperfine tensor are [a − T(1 + δ), a − T(1 − δ), a + 2T] and the rhombicity parameter δ ranges from 0 (axial tensor) to 1 (rhombic tensor). 2c has a very small isotropic constant, thus being represented with only a single cross-ridge symmetric about the diagonal in the FdxB spectra, and points of cross-ridge overlap producing the 1c+2c peaks in the stacked spectra are indicated with orange circles (right). For ARF (B), the overall experimental spectra showing the intense “arc-shaped” 13C crossridges off the antidiagonal line were successfully reproduced by EasySpin simulations with two anisotropic 13Cβ(Cys) tensors of similar low rhombicities, 1c (a = 0.8 ± 0.2 MHz; T = 1.3 ± 0.1 MHz; δ = 0.1 ± 0.1; Euler angles α = −35 ± 10°, β = −59 ± 9°, and γ = 89 ± 33°) and 2c (a = 0.4 ± 0.2 MHz; T = 1.3 ± 0.1 MHz; δ = 0.2 ± 0.1; Euler angles α = 25 ± 22°, β = 53 ± 6°, and γ = 0 ± 14°), where the heavy overlap of 1c and 2c results in peaks (orange circles) that can be identified as only 1c+2c (right). Experimental parameters: magnetic fields of 341.5 mT (g||), 347.0 mT (gint), and 355.7 mT (g⊥); a microwave frequency of 9.646 GHz; and a temperature of 20 K for the FdxB spectra (A) and magnetic fields of 344.0 mT, (gz), 364.0 mT (gy), and 386.2 mT (gx); a microwave frequency of 9.696 GHz; and a temperature of 10 K for the ARF spectra (B). This figure was prepared by Dr. Alexander Taguchi.
For the reduced [2Fe-2S] cluster in P. putida FdxB with a complete cysteinyl coordination (gmax (g||) = 2.020, gint = 1.936, gmin = 1.934), 13C HYSCORE spectra showed four 13Cβ-(Cys) cross-features resolved by optimized EasySpin simulations, with two anisotropically elongated cross-ridges from the Cys86 (1c) and Cys50 (2c) ligands coordinated at the non-reduced Fe3+ site, and two additional components at low contour levels with weak anisotropy (3c and 4c) from Cys41 and Cys47 at the Fe2+ site (Fig. 9A). At the Fe3+ site of the reduced cluster, the magnitude of the electron spin population at Cys50 13Cβ is significantly smaller than that at Cys86 13Cβ in FdxB.
The 13C HYSCORE spectra of reduced archaeal Rieske-type [2Fe-2S](His)2(Cys)2 ferredoxin (ARF, gz = 2.022, gy = 1.901, gx = 1.804) from Sulfolobus solfataricus strain P1 showed only elongated 13Cβ(Cys) cross-ridges that are shifted off the antidiagonal dashed line defined by να + νβ = 2ν13C (Fig. 9B); these cross-features 1c and 2c, characteristic of 13C tensors with significant hyperfine anisotropy, are similar to those observed for the cross-ridges 1c and 2c in the FdxB spectra (Fig. 9A), but have unresolved individual contributions from the Cys42 and Cys61 ligands coordinated at the Fe3+ site (Fig. 9B), owing to the relatively similar magnitudes of the electron spin populations at Cys42 13Cβ and Cys61 13Cβ in reduced ARF.
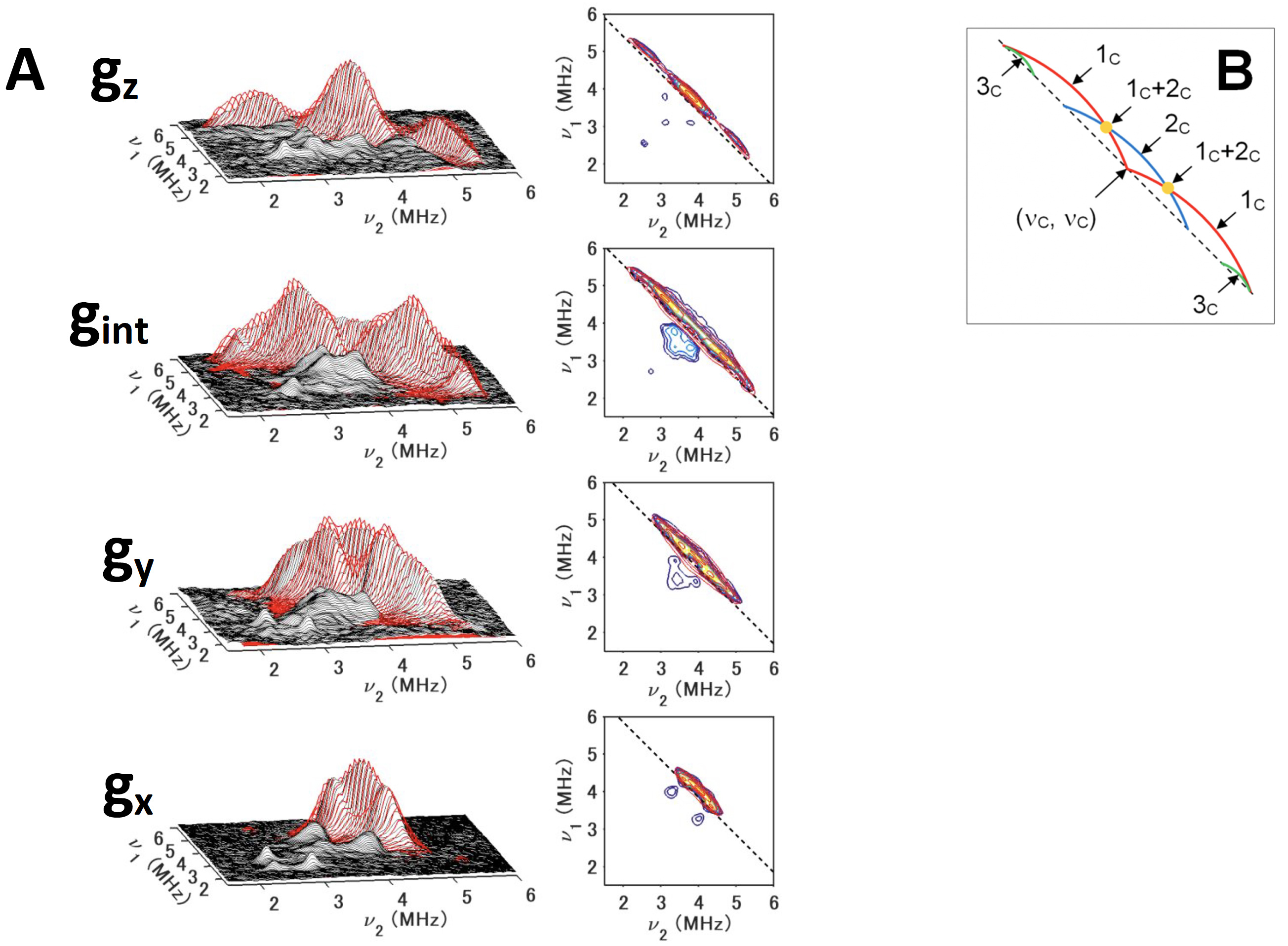
Figure 10. 13Cβ(Cys) HYSCORE spectra of 13Cβ(Cys)-labeled, reduced TthNEET in the 14N (natural abundance)-protein background, measured at gz (345,8 mT) (top), gint (352.8 mT) (first middle) , gy (359.8 mT) (second middle), and gx (366.8 mT) (bottom), shown in three-dimensionally stacked (left) and contour (right) representations, respectively (A), and schematic diagrams of the powder 13C HYSCORE spectra (B) for the simulated cross-ridge line shapes (the schematic figures are not drawn to scale, and the cross-ridge curvature is exaggerated for the sake of clarity). The spectral simulations are colored red overlaying the experimental 13C HYSCORE spectra in the (++) quadrant. In the contour representations, the dashed lines along the antidiagonal are defined by να + νβ = 2ν(13C). In the optimized EasySpin simulations of the TthNEET spectra (A), the inclusion of some rhombicity (δ= 0.2) into the 13C hyperfine tensors for 1c and 2c was found to be necessary to model the experimental HYSCORE cross-ridge shapes. For 3c, the spectral intensity was too low for a meaningful least-squares optimization. Therefore, an axial hyperfine tensor was assumed (δ= 0), and the Euler angles were adjusted manually by eye. This figure was prepared by Dr. Alexander Taguchi.
Out of all three model proteins, TthNEEET, a thermophile mitoNEET [2Fe-2S](His)1(Cys)3 protein homolog from Thermus thermophilus strain HB8, exhibits the largest asymmetry in the ligand type arrangement, with two Cys ligands at the nonreducible Fe3+ site and one Cys ligand at the reduced Fe2+ site. The 13Cβ-Cys HYSCORE spectra show two anisotropic cross-ridges (1c and 2c) and a third isotropic cross-peak 3c at low contour levels (Fig. 10). An important advantage of the two-dimensional pulsed EPR spectroscopic approach is the resolution of cross-ridges from nuclei with different degrees of hyperfine anisotropy. These features would otherwise be masked by more intensive ridges in one-dimensional spectra.
A full analysis of the 13Cβ(Cys) HYSCORE spectra acquired at multiple orientations spanning the full g-tensor for each protein was performed, in conjunction with spectral simulations performed with EasySpin v5.0.9. The 13C hyperfine tensors are grouped by their value of T, where 1c and 2c exhibit a high degree of anisotropy (1.2−1.4 MHz) and 3c and 4c are less anisotropic (0.3−0.5 MHz) (Table 1). The range of these 13Cβ hyperfine anisotropies can be explained by the point−dipole model for the magnetic interaction between the 13C nucleus and the Fe spin density. By modeling the Fe3+ and Fe2+ spin populations with the normalized coefficients DS determined in similar protein systems (+1.60 to +1.85 for Fe3+ and −0.55 to −1.24 for Fe2+), the dipolar contribution to the anisotropic constant for a coordinated Cys-Cβ is estimated to be 0.9−1.0 MHz for the Fe3+ ligands and 0.3−0.7 MHz for the Fe2+ ligands. Comparison of these two scenarios with the values of T in Table 1 allows the clear assignment of 1c and 2c to Fe3+ ligands, and 3c and 4c to Fe2+ ligands. For TthNEET, 3c is immediately assignable to Cys48 at the Fe2+ site (Fig. 11).
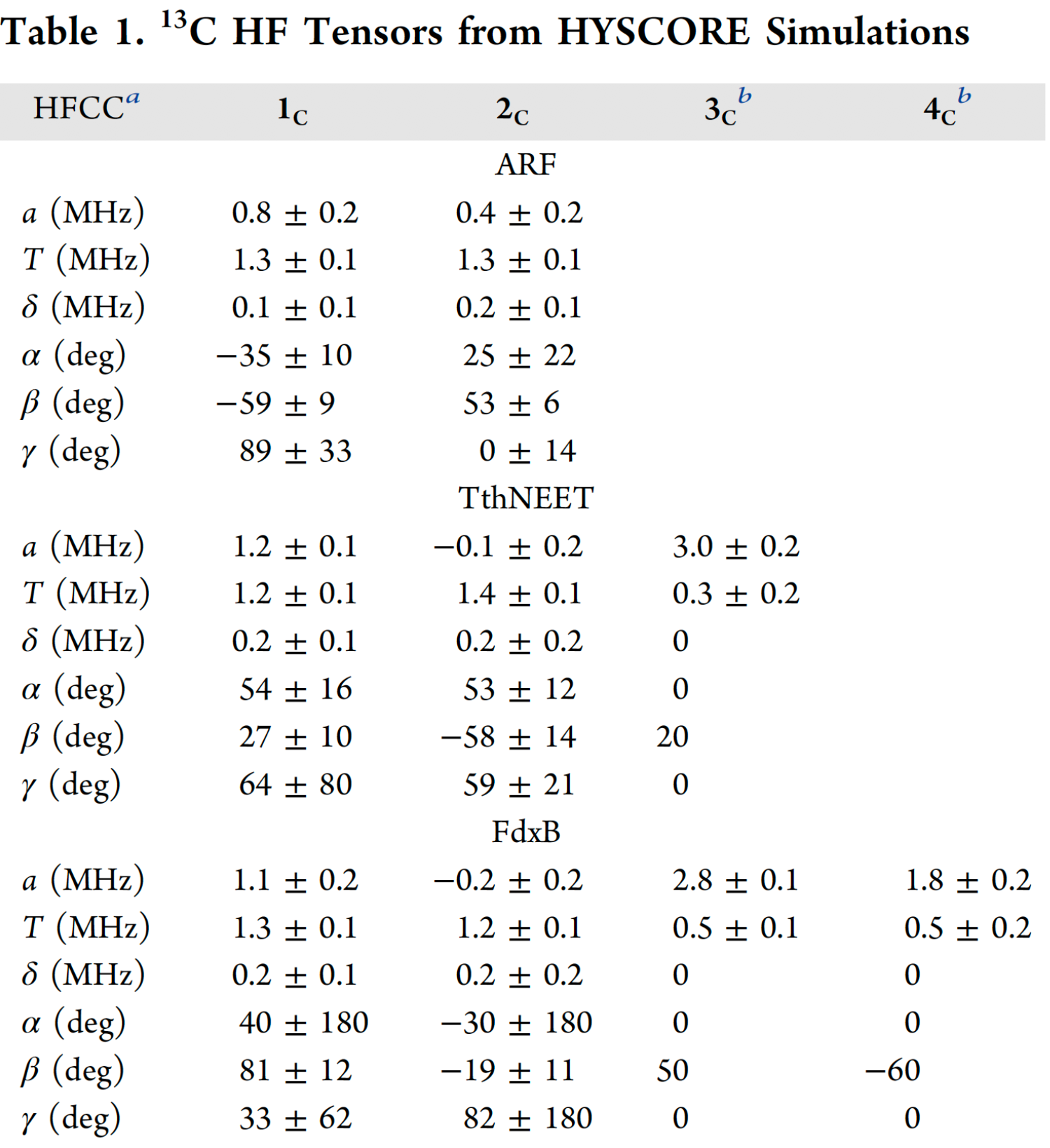
a Hyperfine (HF) coupling constants. a, T, and δ are the isotropic, anisotropic, and rhombic constants, respectively. Euler angles α, β, and γ describe the rotations that bring the g-tensor into the hyperfine tensor eigenframe. b Due to the low intensity of 3c and 4c, axial hyperfine tensors were assumed (δ= 0) and the Euler angles were adjusted manually. The errors reported for a and T were thus estimated by eye.
For the assignments of the 13C tensors to particular nuclei in a residue-specific manner, an analysis of the simulated Euler angles (α, β, γ) within the gtensor reference frame must be considered. Only for FdxB has the g-tensor orientation been determined, and therefore, the discussion will be limited to this system. For the point−dipole interaction between the 13Cβ and its nearest iron, the A∥ component of the HF tensor is expected to lie along the line connecting the two atoms. The Euler angle β (angle between A∥ and gz) predicted from the FdxB structure (3AH7.pdb) is 11° for Cys50 and 82° for Cys86. These values only agree with the assignment of 1c to Cys86 and 2c to Cys50 (Table 1 and Fig. 11). The Fe2+ coordinated cysteines are not considered here, because the Euler angles for these 13Cβ(Cys) tensors (3c and 4c) could not be accurately determined.
The 13Cβ(Cys) isotropic hyperfine couplings in Table 1 are directly related to the degree of unpaired spin delocalization onto the cysteine ligands of the biological [2Fe-2S](His)n(Cys)4-n (n = 0, 1, 2) cluster. After integrating these results with previously reported hyperfine couplings for the remote nitrogens of the histidine ligands, a detailed map of the unpaired s-spin density distribution in the immediate cluster environment relevant to biological electron transfer is constructed (Fig. 11). Our results identified the highly asymmetric pattern of the spin distribution in all three systems, with a consistent trend where approximately five-fold more electron spin density is transferred onto the β-carbons of the Fe2+-bound cysteine ligands than those of Fe3+, despite the lower S = 2 spin of Fe2+ than S = 5/2 of Fe3+. In the case of TthNEET, the Fe2+/Fe3+ ligands are asymmetrically positioned with respect to the reduced cluster (θ of +9° for Cys37, −90° for Cys39, and −20° for Cys48), and as a result the pattern of spin transfer to the Fe3+ cysteine ligands is highly asymmetric, like the case for FdxB (Fig. 11). The 13Cβ spin population at the Fe2+ cysteine in TthNEET was found to be almost 7 times larger than the average value at the cysteines of the Fe3+ site, similar to what is observed for FdxB. These results suggest that the local structural asymmetry enforced by the surrounding polypeptide chain, rather than the types of the cluster ligands, regulates the electron spin distribution across Cβ(Cys) nuclei at the reduced [2Fe-2S] cluster site.
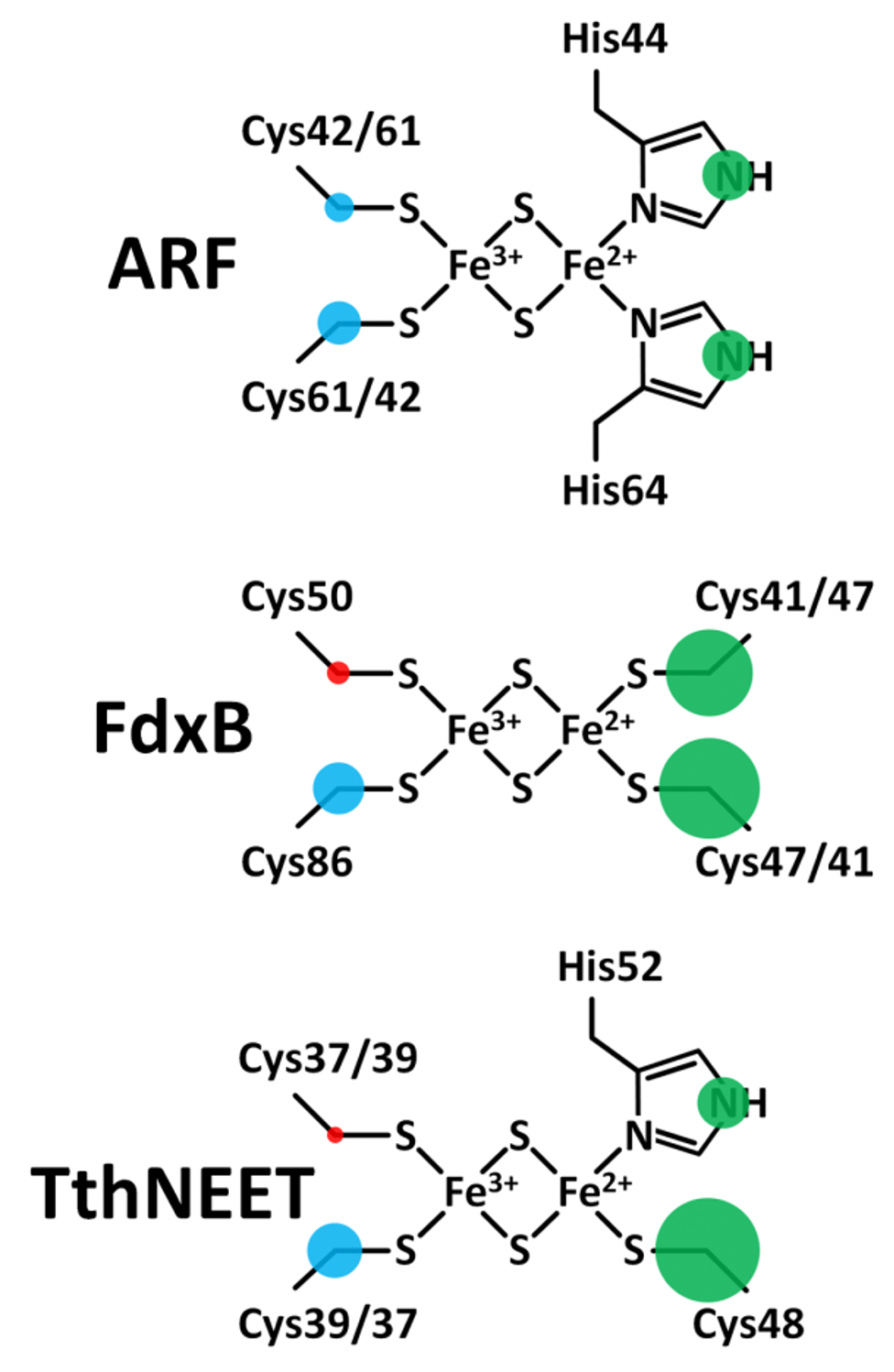
Figure 11. Maps of the outer-shell s-orbital electron spin density distribution onto the ligands of the reduced ARF (top), FdxB (middle), and TthNEET (bottom) clusters. Positive and negative electron spin is shown in blue and red, respectively. Green indicates the sign is unknown. The areas of the circles show the relative spin populations. This figure was prepared by Dr. Alexander Taguchi.
The fundamental redox chemical properties of the biological Fe-S cluster are primarily defined by the covalency of the Fe−ligand interactions. The present 13Cβ(Cys) HYSCORE analysis of the reduced [2Fe-2S](His)n(Cys)4‑n (n = 0, 1, 2) proteins successfully resolves the covalent interactions of the cysteine ligands with the same iron for the first time. The electron spin population on Cβ(Cys) corresponds to the degree of spin delocalization via orbital overlap, and should therefore provide a relative measure of the Sγ(Cys)-Fe covalencies. In ARF, the electron spin populations on 13Cβ of Cys42 and Cys61 were found to differ by only 0.8 X 10^−4, suggesting slightly different covalencies of the two Sγ(Cys)−Fe bonds. For the Fe3+ ligands of FdxB and TthNEET, where the ligand placement relative to the cluster is asymmetrical, the differences in spin populations are greater at 2.3 X 10^−4 and 2.4 X 10^−4, respectively. A similar situation applies to the Fe2+ ligands of FdxB where the difference in Cβ(Cys) spin densities is 2.7 X 10^−4. For the Fe3+ ligands of FdxB, the greater spin transfer to the Cys86-Cβ would suggest that Cys86 has the stronger of the two Sγ(Cys)−Fe covalent interactions; a similar residue-specific analysis for the Fe3+ ligands of TthNEET would first require an assignment of the hyperfine tensors to specific nuclei. The preferred delocalization of the electron spin along the Fe2+ site ligands for all clusters studied (Fig. 11) correlates well with electron transfer directionality of these proteins; redox chemistry is exclusively catalyzed at the reducible Fe2+/Fe3+ site of the cluster, maximizing the electronic coupling with redox partners.
*One can follow the link to read the original article of this work.
up
The 1.80-Å structure of TthNEET0026
The 1.70-Å structure of Caenorhabditis elegans CISD-1/mitoNEET, a KLP-17 tail domain homologue
CISD-1/mitoNEET is a homo-dimeric outer mitochondrial membrane protein harboring unique [2Fe-2S] clusters coordinated by three cysteines and one histidine, thus belonging to the CDGSH Fe-S (iron sulfur) containing protein family. Human isoforms of this protein family include homo-dimeric CISD-2/Miner1/NAF1, localized mainly in the endoplasmic reticulum membrane and having significant structural similarity to CISD-1/mitoNEET, and monomeric CISD-3/Miner2/MiNT, located at the mitochondrial inner membrane and harboring two oxygen-labile [2Fe-2S] clusters per single polypeptide chain. All of these Fe-S clusters are redox active, but substantial differences in their oxygen sensitivities across different isoforms imply some differences in their physiological functions in regulating mitochondrial metabolism, morphology and dynamics. Gain and loss of function studies have shown that the mammalian CISD-1/CISD-2/CISD-3 isoforms play critical roles in oxidative stress and the etiology of multiple disease states including diabetes, breast cancer, and neurodegenerative diseases such as Parkinson’s disease.
The CISD/NEET protein homologues have been found in a variety of phylogenetically distantly related organisms. In free-living transparent nematode Caenorhabditis elegans, which serves as a simple multicellular model eukaryotic system, one cisd-1 homologue (annotated as W02B12.15) and two cisd-3 homologues (namely cisd-3.1 and cisd-3.2) have been detected: the cisd-1 gene putatively encodes two alternative protein isoforms CISD-1a and CISD-1b, with lengths of 103 or 134 amino acids, respectively, depending on the choice of initiation codon, whereas no cisd-2 homologue was detected. Interestingly, C. elegans CISD-1/mitoNEET is shown to play an important role in mitochondrial health and bioenergetics, as seen in mammalian cells, and loss of CISD-1/mitoNEET in C. elegans has led to an increase in reactive oxygen species (ROS), a decrease in ATP production, and changes in the mitochondrial ultrastructure in larval stages with a higher number of hyperfused mitochondria indicative of its potential role in the mitophagy process. This study aims to determine the crystal structure of C. elegans CISD-1/mitoNEET.
The protein sequence of C. elegans CISD-1a/mitoNEET homologue was identified from WormBase (W02B12.15). In efforts to unveil the structural details of this worm protein, a portion of the W02B12.15 cDNA clone coding for the water-soluble domain (residues 32-103) of C. elegans mitoNEET/CISD-1a was amplified by polymerase chain reaction (PCR) with sets of the PCR primers which included 21 extra bases (coding for an engineered Ser-Gly linker and Lys-Lys-Phe-Tyr-Ala sequence from the Rattus norvegicus (rat) heart mitoNEET/CISD-1 sequence) immediately upstream of the codon for conserved Lys32 of the target gene: the engineering of these extra amino acid sequences significantly improved heterologous overexpression of the reddish recombinant protein amenable for the subsequent structural studies. The amplified PCR product was digested with NheI and XhoI, and then inserted into the NheI/XhoI site of a pET28a vector. The pET28aCENEET- SD2 expression vector was transformed into the host strain, E. coli CodonPlus(DE3)-RIL (Stratagene), and used for the following sample preparations.
For preparation of the C. elegans mitoNEET/CISD-1a soluble domain sample, the transformants were grown overnight at 25 °C in a 3 L culture of LB medium containing 50 𝜇g/mL kanamycin and 0.2 mM FeCl3 (Wako Pure Chemicals, Tokyo, Japan), and the recombinant holoprotein was overproduced with 1 mM isopropyl 𝛽-D-thiogalactopyranoside for 19 h at 25 °C. The cells were pelleted by centrifugation, and the recombinant C. elegans mitoNEET (residues 32-103, see below) having a hexa-histidine-tag plus a thrombin cleavage site and an engineered extra linker sequence was purified essentially as reported previously for archaeal Rieske [2Fe-2S] proteins, except that the entire purification was performed at 4 °C using buffers adjusted at pH 8.0 and that the heat treatment of the crude cell lysate was omitted. After proteolytic removal of the hexa-histidine tag from the purified recombinant protein for 16–22 h at 4 °C using a thrombin cleavage capture kit (Novagen) according to the manufacturer’s instructions, the sample was further purified by gel-filtration chromatography (Sephadex G-75; Amersham Pharmacia Biotech), eluted at room temperature with 10 mM HEPES-NaOH buffer, pH 8.0, containing 500 mM NaCl, and then concentrated with Centriprep-10 and Microcon-YM10 apparatuses (Amicon) to ∼4 mM (per protomer), rapidly frozen in liquid nitrogen, and stored at -80 °C until use. The resulting purified protein has the following deduced amino acid sequence: GSHMASSGKKFYA 32KFGQRSARCN YKIQLDSNKI VDTVDIEDIG EKKAFCRCWK SEKWPYCDGS HGKHNKETGD NVGPLIVKSE K 103K.
The purified recombinant C. elegans mitoNEET/CISD-1a soluble domain, containing fifteen extra linker residues (GSHMASSGKKFYAKF) at the N-terminus, which was engineered to improve overproduction and facilitate rapid purification and is absent in the CISD-1a sequence (WormBase accession number W02B12.15), was crystallized under oxygenic conditions by the standard hanging drop vapor diffusion technique. The final conditions were droplets consisting of 1–2 𝜇l of purified protein solution and 1-2 𝜇l of the reservoir solution (100 mM CHES-NaOH, pH 9.5, and 30%(w/v) PEG 3000 (Emerald Biostructures Wizard Classic 1, #41), to which 5%(v/v) of 1 M Bicine, pH 8.5, was added (i.e., 95 mM CHES-NaOH, 50 mM Bicine, 28.5% (w/v) PEG 3000 in the final concentrations)) in the protein well equilibrated against the reservoir solution. Reddish single-crystals (see Fig. 13, inset) were obtained in 1-3 weeks at 4 °C (under oxygenic conditions without any reducing reagents). They were transferred to a cryoprotective solution containing 32.5%(w/v) PEG3000 (Rigaku, Ltd) and frozen in liquid nitrogen. X-ray diffraction data were collected from flash-frozen, small crystals at the SPring-8 beamline BL41XU using a beam size of 25(V) x 9(H) (𝜇m^2) and PILATUS3 6M detector with a detector distance of 250 mm. The data collection was performed at a wavelength of 1.0 Å with a total rotation range of 105° and rotation step of 0.5°/frame. The data was processed up to 1.70-Å resolution by using the XDS package. The crystal belongs to the tetragonal space group P41212, with unit cell parameters a = b = 37.798 Å and c = 82.260 Å. There is a single protein molecule per asymmetric unit.
The residue number for the refined mitoNEET structure quoted here refers to that of the C. elegans CISD-1a isoform in WormBase (W02B12.15). The refined structural model includes the CISD-1a residues 38-100, with Cys40 forming a possible reversible disulfide bond with the SA𝛾-SB𝛾 bond length of 2.5 Å and CA𝛽-SA𝛾-SB𝛾-CB𝛽 torsion angle of +96°(right-handed) at the dimer interface, one [2Fe-2S] cluster and 19 water molecules, and does not contain the engineered extra linker residues (GSHMASSGKKFYAKF) from the pET28aCENEET-SD2 expression vector which are disordered and not resolved in the electron density. The final R factor and Rfree factor were 0.169 and 0.190, respectively. The final structure was deposited in the Protein Data Bank (PDB) with the accession code 7YVZ.
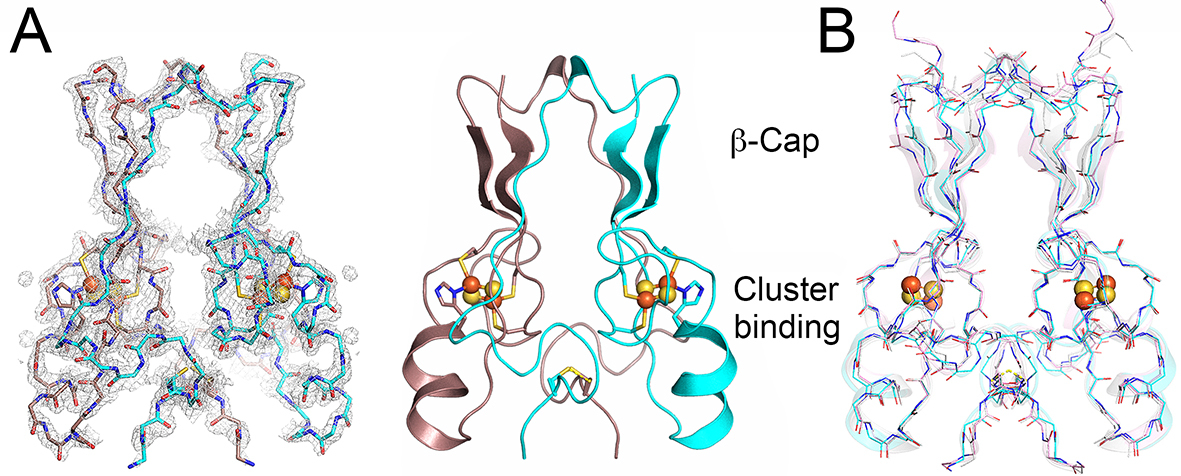
Figure 12. Overall structure and domain topology of C. elegans CISD-1/mitoNEET homodimer (W02B12.15) refined at 1.70-Å resolution (PDB code 7YVZ). (A) The backbone tracing (left) and the cartoon representation (right) of the C. elegans CISD-1/mitoNEET soluble domain. Also shown is the observed 2Fo-Fc electron density map (left, gray) contoured at 1.5 𝜎. Protomers colored in dark pink and cyan, iron in red, sulfur in yellow; (B) The backbone superposition of C. elegans CISD-1/mitoNEET (cyan) on human CISD1/mitoNEET (PDB code 2QH7, gray) and human CISD2/Miner1 (PDB code 3FNV, pink).
Primordial CISD-1/mitoNEET of C. elegans, encoded by the W02B12.15 gene, is a mitochondrial outer membrane protein whose X-ray crystal structure has not been reported previously. The 1.70-Å structure of the soluble-domain of the worm CISD-1 successfully refined to an R factor of 16.9% (Rfree = 19.0%) in this study reveals a tightly packed homo-dimeric structure in the lattice, with each protomer related via a 180° rotation along the crystallographic dyad axis (Fig. 12). Each protomer is folded into two domains, namely, the 𝛽-cap domain and the cluster-binding domain, respectively (Fig. 12A). The 𝛽-cap domain of C. elegans CISD-1 consists of two antiparallel 𝛽-strands and a 310helix, but does not contain the third parallel 𝛽-strand coming from the other protomer found in the closely related, “NEET ”fold structures of human CISD-1/mitoNEET and CISD-2/Miner1/NAF1 (Fig. 12A,B; see also Fig. 15A, top). Superposition of these three NEET protein backbone structures showed key variations around the 310 helix and adjacent loop regions in the 𝛽-cap domain of C. elegans CISD-1, resulting in a backbone conformational shift unfavorable for forming the third parallel 𝛽-strand at the corresponding positions, while the cluster-binding domain and the [2Fe-2S] cluster core of the protomers are remarkably superimposable (Fig. 12B). Structure prediction of C. elegans CISD-1 using ColabFold that combines the fast homology search of MM-seqs2 with AlphaFold2 or RoseTTAFold, also indicated the presence of two anti-parallel 𝛽-strands in the 𝛽-cap domain of the predicted model, being in line with the refined structure (see Fig. 15).
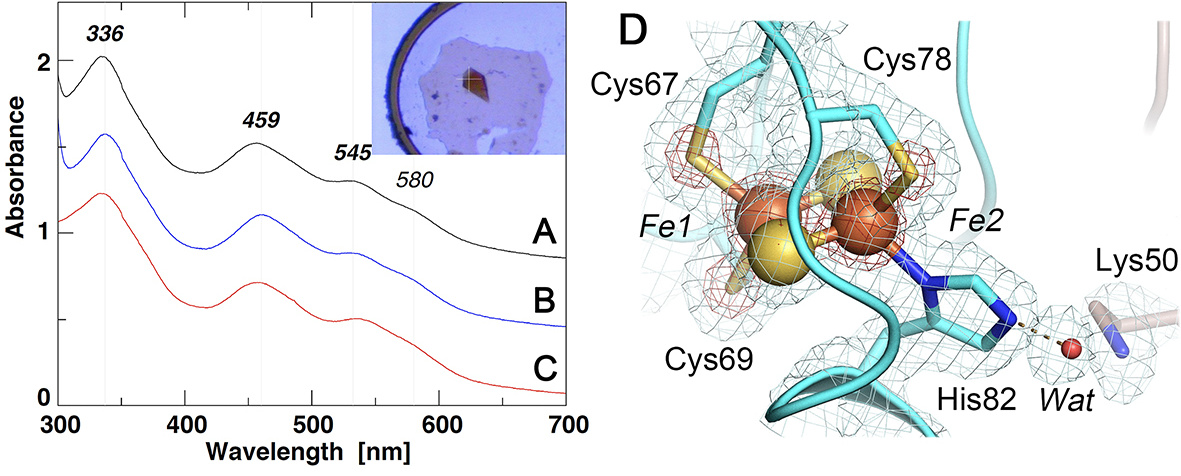
Figure 13. UV-visible absorption spectra of the selected NEET proteins (A-C) and an expanded view of the [2Fe-2S] cluster site of C. elegans CISD-1/mitoNEET (D). (A-C) UV-visible absorption spectra of rat CISD1/mitoNEET (A), C. elegans CISD-1/mitoNEET (inset, its single crystal) (B), and a thermophile homologue, TthNEET, from Thermus thermophilus HB8 (C) (shown with an offset interval of + 0.4 absorbance unit for clarity); (D) An expanded view of the [2Fe-2S] cluster site of C. elegans CISD- 1/mitoNEET together with the observed 2Fo-Fc electron density map contoured at 1.0 𝜎 (gray) and 5.0 𝜎 (red).
The cluster-binding domain of C. elegans CISD-1 consists of a cluster-binding loop with the consensus CXC-//-CDGSH motif, followed by a single 𝛼-helix, and harbors the NEET-type [2Fe-2S] cluster coordinated by one histidine and three cysteine residues, exhibiting the characteristic UV–visible absorption spectra (Fig. 13, 14A). The [2Fe-2S] cluster of C. elegans CISD-1 soluble domain is significantly more stable than regular protein-bound [4Fe-4S] clusters, as evident by the well-defined electron density for the [2Fe-2S] cluster core (Fig. 13D), even after weeks of the hanging drop vapor diffusion setups for crystallization under oxygenic conditions. In the homo-dimeric structure in the lattice, the two [2Fe-2S] clusters are ∼14.6 Å apart as measured by the closest, innermost Fe1-Fe1 distance (coordinated by Cys67 and Cys69) and ∼17.8 Å by the outermost Fe2-Fe2 distance (coordinated by Cys78 and His82), suggesting that a rapid interprotomer electron transfer equilibrium between them would occur. Interestingly, the bridging sulfide (S1) of the cluster engages in N-H…S hydrogen-bonding interaction with Arg68N 𝛼, whose guanidium group also interacts with the mainchain oxygen atoms of Cys67 (one of the ligand residues to the innermost Fe1 site) and Pro76 (in a cis conformation) of the other protomer via a structurally well-conserved, interior water molecule (Wat) by the interprotomer hydrogen bond network (Fig. 14B). Pro56, along with Phe66, Trp70, Trp75, Leu96, also constitute the hydrophobic core at the homodimer interface. Our previous pulsed electron paramagnetic resonance (EPR) analyses clearly showed that the outermost Fe2 site of the [2Fe-2S] cluster is reduced upon chemical reduction in rat mitoNEET and its thermophile homologue. In this regard, although C. elegans CISD-1 was crystallized under oxygenic conditions in the absence of any reducing reagent, exposure of the resulting microcrystals to very high flux X-radiation at the SPring-8 beamline BL41XU resulted in conversion to the photo-reduced form, so the outcome probably represents a mixture of redox states: it should be kept in mind that redox state is actually very difficult to ascertain solely from crystal structures, and that complications due to X-ray photoreduction and methods to generate a specific redox state in crystals can often occur as well.
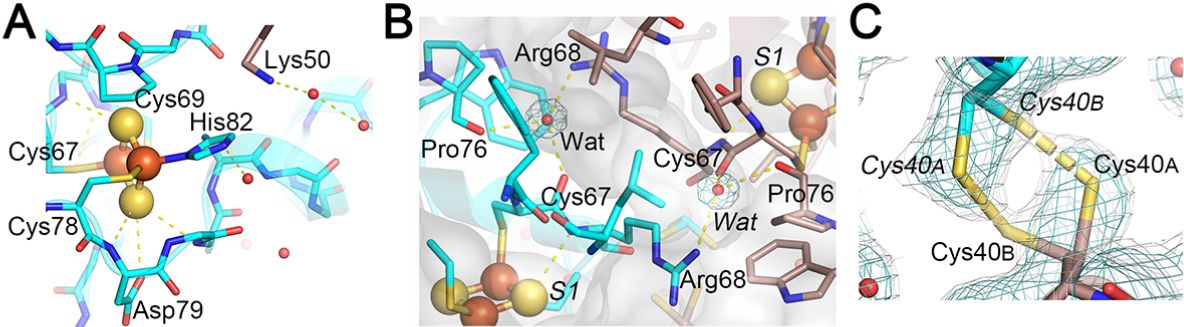
Figure 14. Expanded views of the N-H…S hydrogen-bonding interactions around the [2Fe-2S] cluster (A), the interprotomer hydrogen-bonding interactions between the two [2Fe-2S] clusters (B), and a reversible disulfide linkage (right-handed) contributed from Cys40 conformer A from one protomer and conformer B from the other protomer, together with the observed 2Fo-Fc electron density map contoured at 1.0 𝜎 (cyan) and 0.6 𝜎 (gray) (C) in the C. elegans CISD-1/mitoNEET structure. In (C), the electron density map contoured at 0.6 𝜎 (gray) in PyMOL (Schrödinger, LLC) resembles that observed contoured at 1.0 𝜎 in Coot used for the refinement (not shown).
One of the most unique structural features of C. elegans CISD-1 is the presence of a reversible disulfide linkage contributed from Cys40S𝛾 of each protomer, with the SA𝛾-SB𝛾 bond length of 2.5 Å and CA𝛽-SA𝛾-SB𝛾-CB𝛽 torsion angle of +96°(right-handed) at the homo-dimeric interface in the crystal lattice (Fig. 14C). The equivalent cysteine residue is also conserved in the Drosophila melanogaster CISD-2 sequence but not in mammalian CISD1/mitoNEET and CISD2/Miner1/NAF1 homologues (Fig. 15A). Thus, this is the first example of a eukaryotic NEET protein of this kind. Previously, Conlan et al. have reported substantial flexibility in the N-terminal tethering arms of the human CISD1/mitoNEET soluble domain structure, despite little variations in the 𝛽-cap domain and the cluster-binding domain structures. The presence of a reversible disulfide linkage at the interprotomer interface in C. elegans CISD-1 (Fig. 14C) implies a redox-linked control of multiple orientations on the outer mitochondrial membrane that may be functionally important for partner-protein interactions.
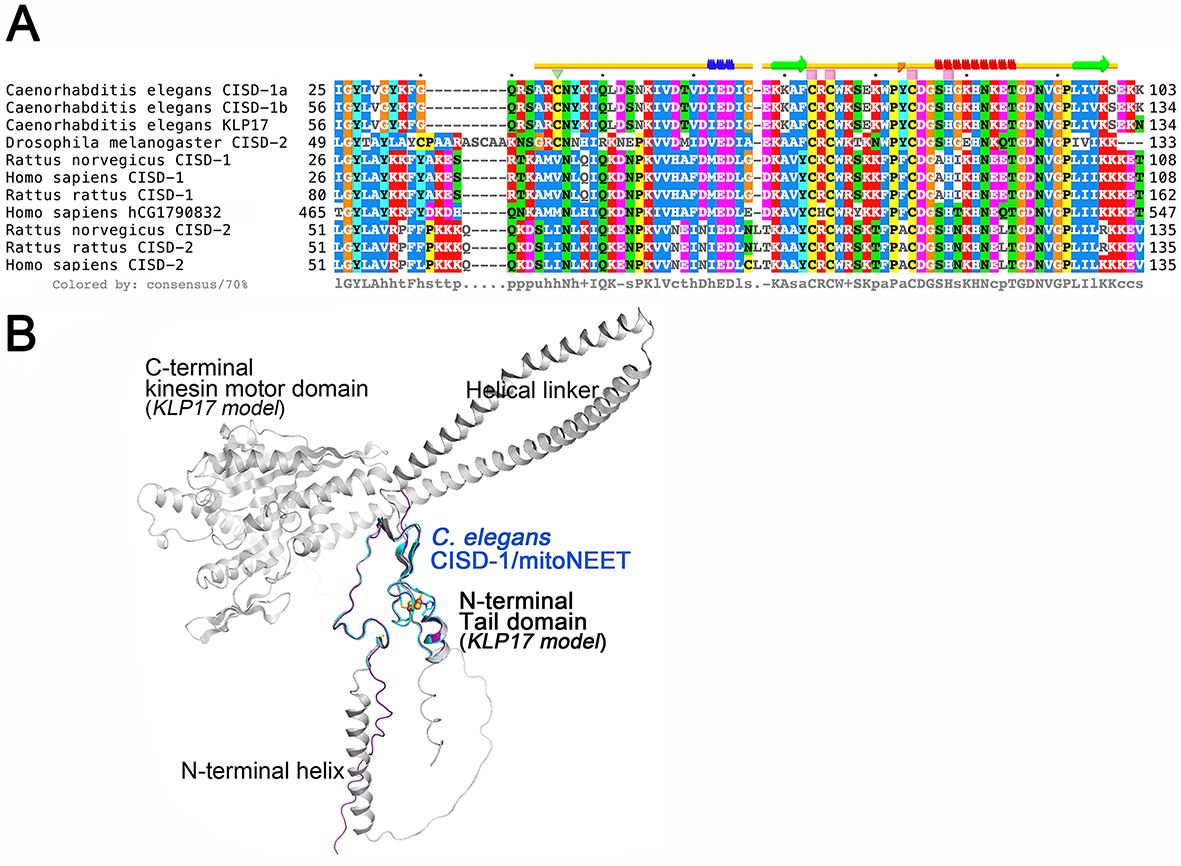
Figure 15. Multiple sequence alignment of selected CISD-1/CISD-2 and related proteins (A) and the superposition of the crystal structure of C. elegans CISD-1/mitoNEET protomer (cyan) on the predicted structural models by ColabFold of the cognate kinesin motor protein KLP17 (gray) and the entire amino acid sequence region of purified C. elegans CISD-1/mitoNEET used for crystallization (purple) (B). (A) Multiple amino acid sequence alignments of the selected proteins were performed by using the EMBL-EBI Clustal Omega server, and the secondary structural elements in the crystal structure of C. elegans CISD-1/mitoNEET (top) were analyzed by using the STRIDE server; (B) Note that the N-terminal tail domain of KLP17 (gray) exhibits the identical structure as found for the crystal structure (cyan) and the ColabFold-predicted structural model (purple) of the C. elegans CISD-1/mitoNEET soluble domain.
Unexpectedly, the NCBI Protein BLAST search against databases of selected eukaryotic species reveals that C. elegans CISD-1 sequence is highly homologous to not only the CISD-1/mitoNEET and CISD- 2/Miner1/NAF1 sequences but also the N-terminal tail domain of the cognate kinesin motor protein KLP17 (W02B12.7 in WormBase, 605 amino acids) and the C-terminal domain of human hCG1790832 gene product of poorly defined function (547 amino acids) (Fig. 15A). This was overlooked in the previous studies presumably because of their substantially larger sizes of the predicted proteins than the CISD1/CISD2 protein orthologues (typically ∼100–150 amino acids). Most strikingly, residues 38–100 covered in the refined structure of C. elegans CISD-1 are completely identical to residues 60-122 in the KLP17 tail domain. Thus, the refined structure of C. elegans CISD-1 soluble domain reported herein also represents that of the homo-dimeric N- terminal tail domain (residues 60-122) of the cognate kinesin motor protein KLP17 (Fig. 15). Structure prediction of the entire C. elegans KLP17 protomer using ColabFold suggests an N-terminal signal sequence plus an 𝛼-helix region, followed by the CISD-1 tail domain for homo-dimerization, which is further connected to the C-terminal globular kinesin motor protein domain with an ATP binding cassette motif, via two long 𝛼-helices linking these two functional domains (Fig. 15B). In the previous studies, Siddiqui and coworkers have reported that the klp-17 transcripts are confined specifically to the cell nucleus during early development, from the one-cell stage of embryogenesis until early larval stages, and that loss of klp-17 via RNAi results in embryonic lethality at the one- or two-cell stage with disorganized mitotic spindles and polyploid nucleus. Our present results indicate that C. elegans KLP17 is a unique member of the C-terminal kinesin motor protein family that contains the N-terminal CISD-1 dimerization tail domain harboring the NEET-type [2Fe-2S](His)1(Cys)3 cluster (Fig. 15) and likely participates in chromosome segregation dynamics in the cell nucleus and germline development. The striking structural similarity of the N-terminal tail domain of KLP17 and CISD-1/mitoNEET (Fig. 15) also implies a possible inter-organellar cross-talk between the cell nucleus and mitochondria by certain redox and/or metabolic status signals in the germline development of C. elegans. Further studies on the worm KLP17 will clarify the structure and physiological function of this new NEET-kinesin motor fusion protein.
The [2Fe-2S](His)1 (Cys)3 cluster-containing mitochondrial outer membrane protein CISD-1/mitoNEET from C. elegans has been proposed to play versatile roles ranging from mitochondrial metabolism and bioenergetics to overall fitness and germline viability. The 1.70-Å structure of the CISD-1 soluble domain shows a tightly packed homo-dimeric structure in the crystal lattice, each protomer having the “NEET” fold structures of human homologues, yet containing a reversible disulfide linkage at the homodimer interface which is apparently conserved in the Drosophila homologues but not in mammalian orthologues. Surprisingly, the refined CISD-1 structure also appears to represent that of the N-terminal tail domain of the cognate kinesin motor protein, KLP-17, involved in chromosome segregation dynamics and germline development of the worms.
*One can follow the link to read the original article of this work.
 HOME
HOME
 Research Top
Research Top E. coli auxotrophic expression strains
E. coli auxotrophic expression strains 古細菌 Archaea | Sulfurisphaera tokodaii (f. Sulfolobus tokodaii)
古細菌 Archaea | Sulfurisphaera tokodaii (f. Sulfolobus tokodaii) モデル鉄硫黄蛋白質|Fe-S world in Sulfolobus
モデル鉄硫黄蛋白質|Fe-S world in Sulfolobus ISC-like ferredoxin (FdxB) from P. putida
ISC-like ferredoxin (FdxB) from P. putida Microbial "mitoNEET" homologs
Microbial "mitoNEET" homologs Ligand mutagenesis
Ligand mutagenesis ICC Project
ICC Project Publication List
Publication List Go to the Research Top Page
Go to the Research Top Page Go to the ICC Project Details Page
Go to the ICC Project Details Page