日本医科大学、医学部、生化学・分子生物学(代謝・栄養学)所属の 岩崎 俊雄グループでは、好熱菌等を主材料とした各種金属酵素の構造機能進化や、アミノ酸要求性の新規大腸菌発現宿主株作成につき、国内外の様々な研究機関と共同研究を実施しています。
Archaea
The idea of using amino-acid sequences as a tool for molecular phylogeny was first proposed by Francis Crick in 1958. In the 1970s, Carl Woese and coworkers revisited the problem of prokaryotic taxonomy by the molecular biological approach with the small-subunit (SSU) ribosomal RNA (rRNA) sequence as a molecular chronometer. As an essential component of all self-replicating organisms, rRNA shows remarkable sequence conservation, with different parts of the molecule having varying rates of base substitution. Moreover, rRNA is abundant and easy to isolate, which has facilitated phylogenetic analyses of "cultivation-independent" microorganisms.
"Archaea" @ University of Illinois at Urbana-Champaign.
Copyright 2012 Toshio Iwasaki. All Rights Reserved.
The archaeal domain contains microorganisms having the most extraordinary optimal growth conditions, with members flourishing at the extremes of pH, temperature, and salinity. The rRNA tree reveals that the domain Archaea comprises several phyla. Among them, members of Euryarchaeota represent the most diverse group, including all known methanogens and halophiles, in addition to thermophilic and psychrophilic species. Members of Crenarchaeota are renowned as hyperthermophiles, but include the psychrophile Cenarchaeum symbiosum. Of the remaining phyla, Nanoarchaeota has one known member, Nanoarchaeum equitans, and Korarchaeota are indicated only by environmental DNA sequences so far. The positions of these phyla on the rRNA tree are thus rather uncertain [indicated by dashed branches as below].
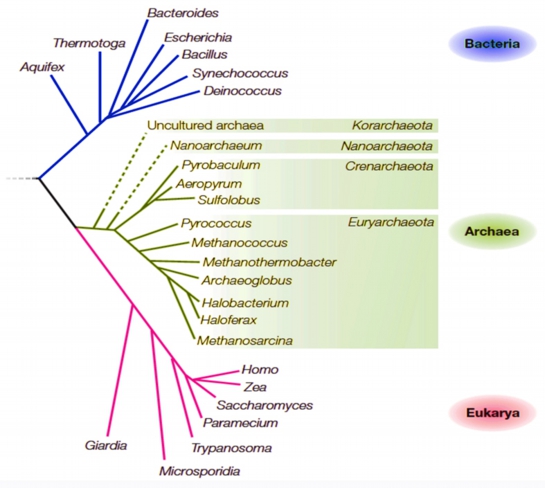
Schematic (proposed) universal phylogenetic tree of life based on 16S rDNA sequences
[adapted from Allers and Mevarech, Nat. Rev. Genetics 6, 58-73 (2005)].
The universal genealogical tree indicates that life diverged first into Bacteria and Archaea. Eukarya then evolved from archaea-like precursor, obtaining mitochondria from alpha-proteobacterial ancestor and chloroplasts from cyanobacterial ancestor. Although genes are passed vertically from a parent organism to its offspring from generation to generation, this vertical inheritance is not the sole key factor that has affected the evolution of organisms. Lateral, or horizontal, gene transfer event, which involves transfer of single genes or whole suites of them across species barriers, is also significant but still rare enough that an overall tree-like structure is maintained. Note that, if there had never been any lateral gene transfer event, all individual gene tree would have the same topology, but this is not the case.
Possible extent of lateral gene transfer events at the root is uncertain, but it would explain how Archaea obtained genes usually found in Bacteria, and vice versa. It also means that "the last universal common ancestor" cannot have been a particular single organismal lineage. One possibility is that it was communal, diverse conglomeration of primitive organisms that genetically formed a web of untree-like links between branches at the root - the three major domains of life, Archaea, Bacteria, and Eukarya, likely arose from an ancestral population of primitive organisms.
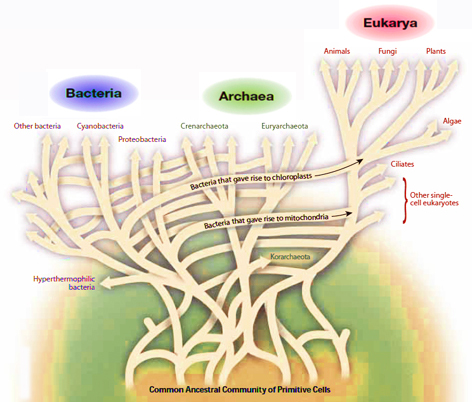
Evolutionary web hypothesis, illustrating tree-like structure at the top of the three domains of life, and an extensive network of untree-like links between branches at the root [adapted from Doolittle WF, Sci. Amer. 282, 90-95 (Feb, 2000)].
More recently, a new comprehensive tree of life that encompasses the total diversity represented by sequenced genomes has been reported by Hug et al., making use of genomes from public databases and over 1,000 newly reconstructed genomes recovered from a variety of environments by metagenomics. While this new view of the tree of life is largely congruent with the traditional SSU rRNA gene sequence-based tree of expected organism groupings at the taxonomic levels, the large number of major lineages without isolated representatives (red dots in the figure shown below), such as the Candidate Phyla Radiation (CPR, many of which are inferred to be symbionts), is highly striking.
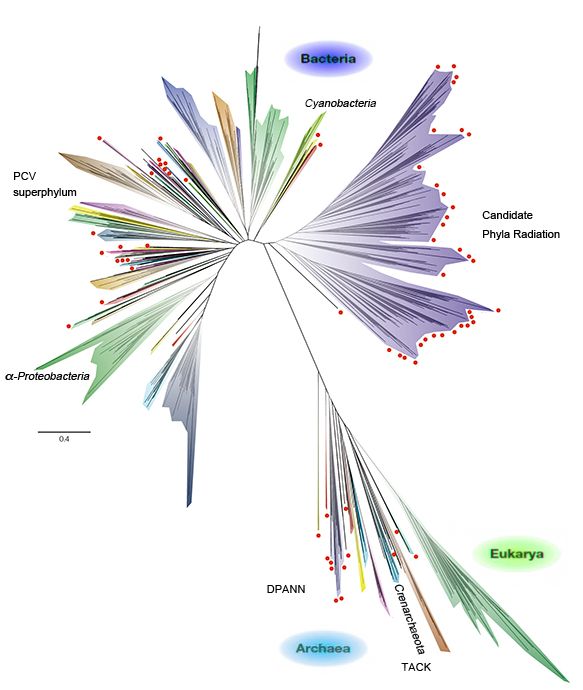
A new schematic view of the tree of life, encompassing the total diversity represented by sequenced genomes [adapted from Hug et al., Nature Microbiology 1, article number: 16048 (2016); see this article for detailed information].
In this new tree, the domain Bacteria includes more major lineages of organisms than the other domains. The domain Archaea is less diverse and less prominant in many ecosystems, and the domain Eukarya exhibits even lower apparent phylogenetic diversity due to their comparatively recent evolution.
Many archaeal species exhibit unique morphological and/or metabolic features [e.g., see Adam et al. (2017) The ISME Journal 11, 2407; Spang et al. (2017) Science 357, 563; Nayfach et al. (2021) Nature Biotechnology 39, 499]. Some of these properties can also be found in the wiki.
up
Sulfurisphaera tokodaii (formerly Sulfolobus tokodaii strain 7)
The archaeal domain contains organisms having the most extraordinary optimal growth conditions, with members flourishing at the extremes of pH, temperature, and salinity. The majority of thermophilic archaea are anaerobic organisms, because oxygen is often scarce in their habitats. The archaeal order Sulfolobales within the Crenarchaeota kingdom of the Archaea domain is composed mainly of the genera Sulfolobus, Acidianus, Metallosphaera, Stygiolobus, Sulfurisphaera, and Sulfurococcus. These thermoacidophilic cocci have been isolated from continental solfataric fields and smoldering mining refuse piles and phylogenetically characterized. Sulfolobus and Metallosphaera are obligate aerobes, while Acidianus and Sulfurisphaera are facultative anaerobes. Stygiolobus is an obligate anaerobe. The taxonomy of the order Sulfolobales based on the early phenotypic properties is somewhat confusing (i.e., it does not always coincide with the inferences derived from the phylogenetic analysis of their 16S rDNA sequences).
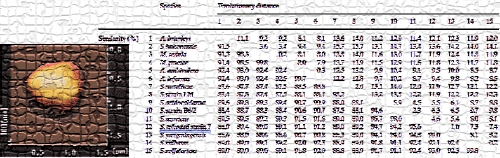
The thermoacidophilic archaeon, Sulfolobus sp. strain 7 sp. nov. (formerly called S. acidocaldarius strain 7), was isolated from the geothermal area of an acidic spa in Beppu Hot Springs (Myoban Spa, "明礬温泉" in Japanese), Kyushu, Japan in the early 1980s by Dr. Tairo Oshima and his colleagues and initially characterized by its phenotypic properties. After continuous cultivation in the Dr. Oshima's lab over 20 years, the taxonomic position of this crenarchaeote was characterized by cloning and sequencing, by phylogenetic analysis of the 16S rRNA gene sequence, by DNA–DNA homology with similar species, and by biochemical characterization of the isolate—we published these results in 2002. This isolate is an obligate aerobe and grows optimally at 80°C and pH 2.5–3 under aerobic and chemoheterotrophic growth conditions by aerobic respiration rather than simple fermentation. In conjunction with the phenotypic properties, the phylogenetic analysis based on the 16S rRNA gene sequence and DNA–DNA hybridization experiments showed that this isolate is related to the described Sulfolobus taxon and should be considered a novel species of the genus (Fig. 1). This isolate is a novel species of the genus Sulfolobus that we named Sulfolobus tokodaii sp. nov. (tou.kou.dye. M. L. gen. adj. tokodaii, pertaining to the Japanese name of the Tokyo Institute of Technology "東工大", where this isolate was initially characterized in the Dr. Oshima's lab). The type strain is strain 7 (JCM 10545, Japan Collection of Microorganims, RIKEN, Japan). More recently, Sulfolobus tokodaii Suzuki et al. 2002 has been reclassified as Sulfurisphaera tokodaii comb. nov. (type strain 7 = JCM 10545 = DSM 16993) by Tsuboi et al. 2018.
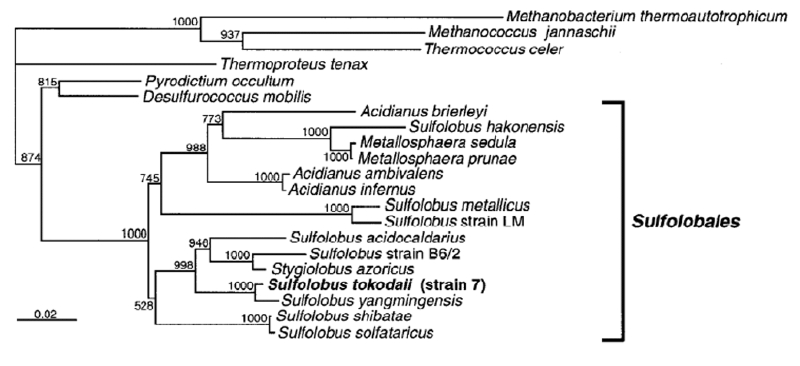
Figure 1. Phylogenetic tree for the Sulfolobales. The tree is derived from the multiple nucleotide sequence alignments of the 16S rRNA genes of the representatives of selected archaea including S. tokodaii strain 7 by the neighbor-joining method. Recently, reclassification of Sulfolobus tokodaii Suzuki et al. 2002 as Sulfurisphaera tokodaii comb. nov. has been reported by Tsuboi et al. 2018.
The S. tokodaii cells are irregular cocci and variable in size (usually 0.5-0.8 micrometers in diameter) and have some flattened and uneven surfaces (Fig. 2). The cell envelope consists of a hexagonal S-layer lattice (spacing approximately 20 nm). Colonies are pale tan, translucent, smooth, and convex, with entire margins.

Figure 2. Atomic force microscope image probing a typical cell shape and uneven surface of S. tokodaii strain 7.
Cells grow chemoheterotrophically on a modified Brock’s basal salts mixture supplied with trypton, yeast extract, casaminoic acid, or amino acids under obligatory aerobic conditions. Best growth occurs on a modified Brock’s basal salts mixture supplied with yeast extract (1 g/L), casaminoic acid (1 g/L), and glucose (1 g/L). Supplements of elemental sulfur are not required for the chemoheterotrophic growth. Poor growth occurs under facultatively chemolithotrophic conditions in the presence of elemental sulfur when supplied with alanine (10 mg/L) or polypeptone (0.1 g/L). No growth occurs in minimal medium amended with a H2/CO2 (90 : 10) headspace. Growth occurs at 70° to 85°C; optimal growth is at 80°C under aerobic and chemoheterotrophic growth conditions. No growth was detected at 65° and 90°C. Growth occurs at pH 2 to 5; optimal growth is at pH 2.5–3. No growth was detected at pH 1 or pH 6. It is an obligately aerobic and facultatively chemoheterotrophic thermoacidophile that grows by aerobic respiration rather than by simple fermentation. Cells contain membrane-bound cytochromes b562, a583, and aa3, with the α-bands at 562 nm, 583 nm, and 603 nm, respectively, as constituents of its aerobic respiratory terminal oxidase supercomplex (a functional fusion of respiratory complexes III and IV) and a soluble zinc-containing ferredoxin as a major cytosolic electron transport protein. No c- and d-type cytochromes are detected spectrophotometrically.
A number of key enzymes involved in the cognate central metabolic pathways and the bioenergetic system have been subjected to extensive biochemical and molecular biological investigation. For example, sequence analysis of the AoA1-/VoV1-type ATP synthase subunits from S. tokodaii strain 7 has provided a clue to the putative evolution of eukaryal species.
The complete genomic DNA sequence of Sulfurisphaera tokodaii strain 7 (f. Sulfolobus tokodaii strain 7) has been analyzed in 2001 by the NITE Biological Resource Center, Japan. The S. tokodaii strain 7 genome is 2,694,756 bp long, and contains approximately 2,900 ORFs, but the functions of much less than half of them are known. The G + C content of the genomic DNA is 32.8 mol%, as calculated from its whole genomic DNA sequence. The type strain is Sulfurisphaera tokodaii comb. nov. (f. Sulfolobus tokodaii, type strain 7 = JCM 10545 = DSM 16993), which was isolated from the Beppu Hot Springs in the continental acidic geothermal area of Kyushu Island, Japan, and it has been deposited in the Japan Collection of Microorganisms as strain JCM 10545.
*One can follow the links to read the original article of this work or learn more about the whole genome project of this organism.
up
 HOME
HOME
 Research Top
Research Top E. coli auxotrophic expression strains
E. coli auxotrophic expression strains 古細菌 Archaea | Sulfurisphaera tokodaii (f. Sulfolobus tokodaii)
古細菌 Archaea | Sulfurisphaera tokodaii (f. Sulfolobus tokodaii) モデル鉄硫黄蛋白質|Fe-S world in Sulfolobus
モデル鉄硫黄蛋白質|Fe-S world in Sulfolobus ISC-like ferredoxin (FdxB) from P. putida
ISC-like ferredoxin (FdxB) from P. putida Microbial "mitoNEET" homologs
Microbial "mitoNEET" homologs Ligand mutagenesis
Ligand mutagenesis ICC Project
ICC Project Publication List
Publication List Go to the Research Top Page
Go to the Research Top Page